Outcome Models
Lucy D’Agostino McGowan
Outcome Model (matching)
Outcome Model (matching)
Application Exercise
10:00 - Get starter files from
https://github.com/sta-779-s25/appex-04-YOUR-GITHUB-HANDLE.git - Fit the propensity score matching model according to our DAG
- Fit the outcome model to the matched data
- Estimate the causal effect
Outcome Model (weighted)
✅ This will get us the point estimate
❌ This will get NOT us the correct confidence intervals
Estimating Uncertainty
- Bootstrap
- Sandwich estimator (on outcome model only)
- Sandwich estimator (on outcome model + propensity score model)
1. Create a function to run your analysis once
(on a sample of your data)
fit_ipw <- function(split, ...) {
.df <- analysis(split)
# fit propensity score model
propensity_model <- glm(
qsmk ~ sex +
race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) +
smokeyrs + I(smokeyrs^2) + exercise + active +
wt71 + I(wt71^2),
family = binomial(),
data = .df
)
# calculate inverse probability weights
.df <- propensity_model |>
augment(type.predict = "response", data = .df) |>
mutate(wts = wt_ate(.fitted, qsmk, exposure_type = "binary"))
# fit correctly bootstrapped ipw model
lm(wt82_71 ~ qsmk, data = .df, weights = wts) |>
tidy()
}2. Use {rsample} to bootstrap our effect
2. Use {rsample} to bootstrap our effect
# Bootstrap sampling with apparent sample
# A tibble: 1,001 × 2
splits id
<list> <chr>
1 <split [1566/583]> Bootstrap0001
2 <split [1566/595]> Bootstrap0002
3 <split [1566/577]> Bootstrap0003
4 <split [1566/572]> Bootstrap0004
5 <split [1566/565]> Bootstrap0005
6 <split [1566/562]> Bootstrap0006
7 <split [1566/555]> Bootstrap0007
8 <split [1566/572]> Bootstrap0008
9 <split [1566/579]> Bootstrap0009
10 <split [1566/570]> Bootstrap0010
# ℹ 991 more rows2. Use {rsample} to bootstrap our effect
2. Use {rsample} to bootstrap our effect
2. Use {rsample} to bootstrap our effect
# Bootstrap sampling with apparent sample
# A tibble: 1,001 × 3
splits id boot_fits
<list> <chr> <list>
1 <split [1566/579]> Bootstrap0001 <tibble [2 × 5]>
2 <split [1566/581]> Bootstrap0002 <tibble [2 × 5]>
3 <split [1566/608]> Bootstrap0003 <tibble [2 × 5]>
4 <split [1566/555]> Bootstrap0004 <tibble [2 × 5]>
5 <split [1566/579]> Bootstrap0005 <tibble [2 × 5]>
6 <split [1566/577]> Bootstrap0006 <tibble [2 × 5]>
7 <split [1566/562]> Bootstrap0007 <tibble [2 × 5]>
8 <split [1566/562]> Bootstrap0008 <tibble [2 × 5]>
9 <split [1566/580]> Bootstrap0009 <tibble [2 × 5]>
10 <split [1566/602]> Bootstrap0010 <tibble [2 × 5]>
# ℹ 991 more rows2. Use {rsample} to bootstrap our effect
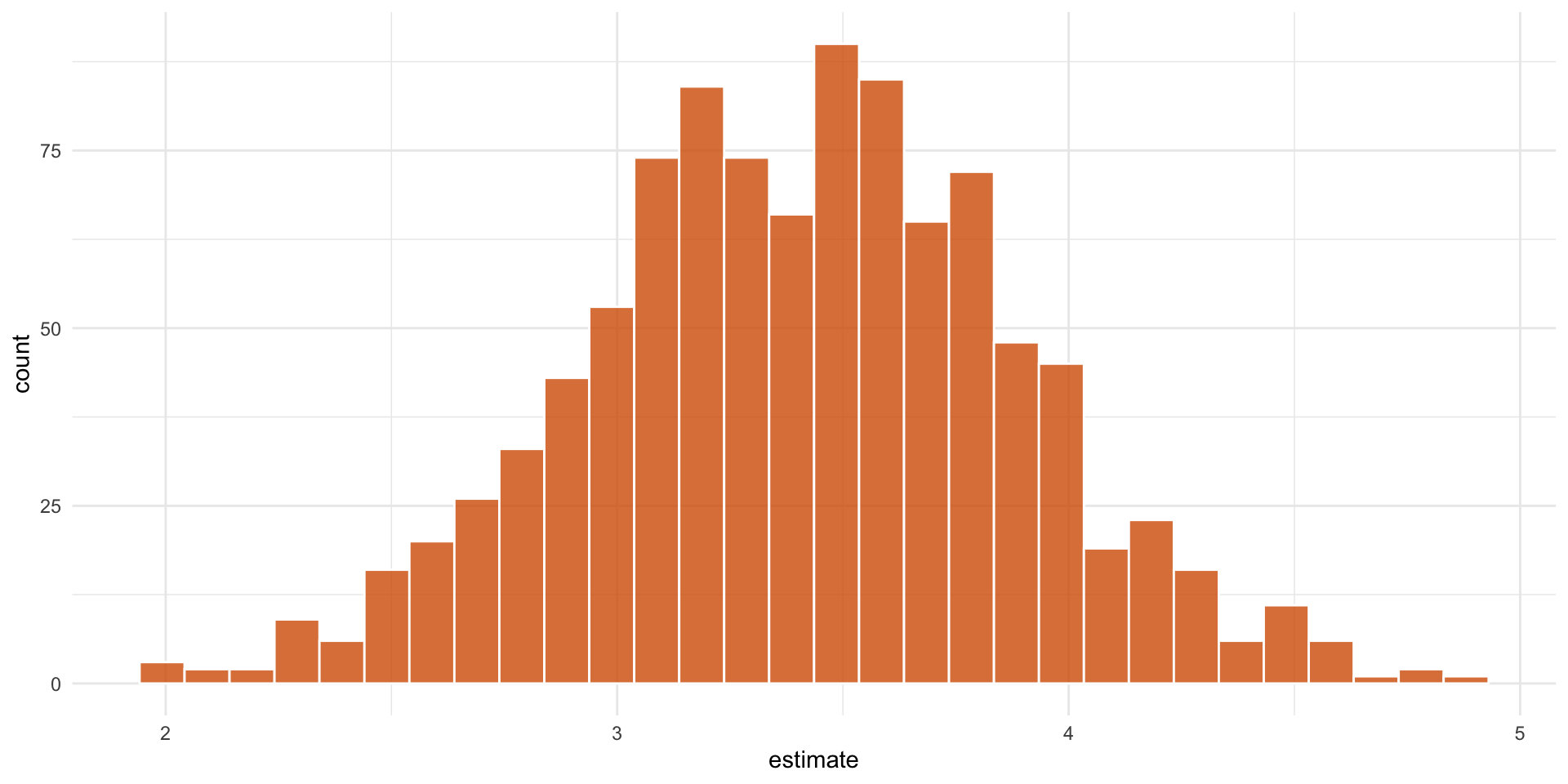
3. Pull out the causal effect
Application Exercise
12:00 Create a function called
ipw_fitthat fits the propensity score model and the weighted outcome model for the effect between the exposure and outcomeUsing the
bootstraps()andint_t()functions to estimate the final effect.
Sandwich estimator
Sandwich Estimator
Sandwich estimator with {survey}
Sandwich estimator with {survey}
Application Exercise
10:00 Use the
sandwichpackage to calcualte the uncertainty for your weighted modelRepeat this using the
surveypackage
Sandwich Estimator with {propensity}
This takes into account that you fit the propensity score model.
Inverse Probability Weight Estimator
Estimand: ATE
Propensity Score Model:
Call: glm(formula = qsmk ~ sex + race + age + I(age^2) + education +
smokeintensity + I(smokeintensity^2) + smokeyrs + I(smokeyrs^2) +
exercise + active + wt71 + I(wt71^2), family = binomial(),
data = nhefs_complete_uc)
Outcome Model:
Call: lm(formula = wt82_71 ~ qsmk, data = nhefs_complete_uc, weights = wts)
Estimates:
estimate std.err z ci.lower ci.upper conf.level p.value
diff 3.4405 0.48723 7.06145 2.4856 4.3955 0.95 1.648e-12 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Sandwich Estimator with {propensity}
This takes into account that you fit the propensity score model.
Application Exercise
10:00 - Use the
{propensity}package to calcualte the uncertainty for your weighted model
Slides by Dr. Lucy D’Agostino McGowan